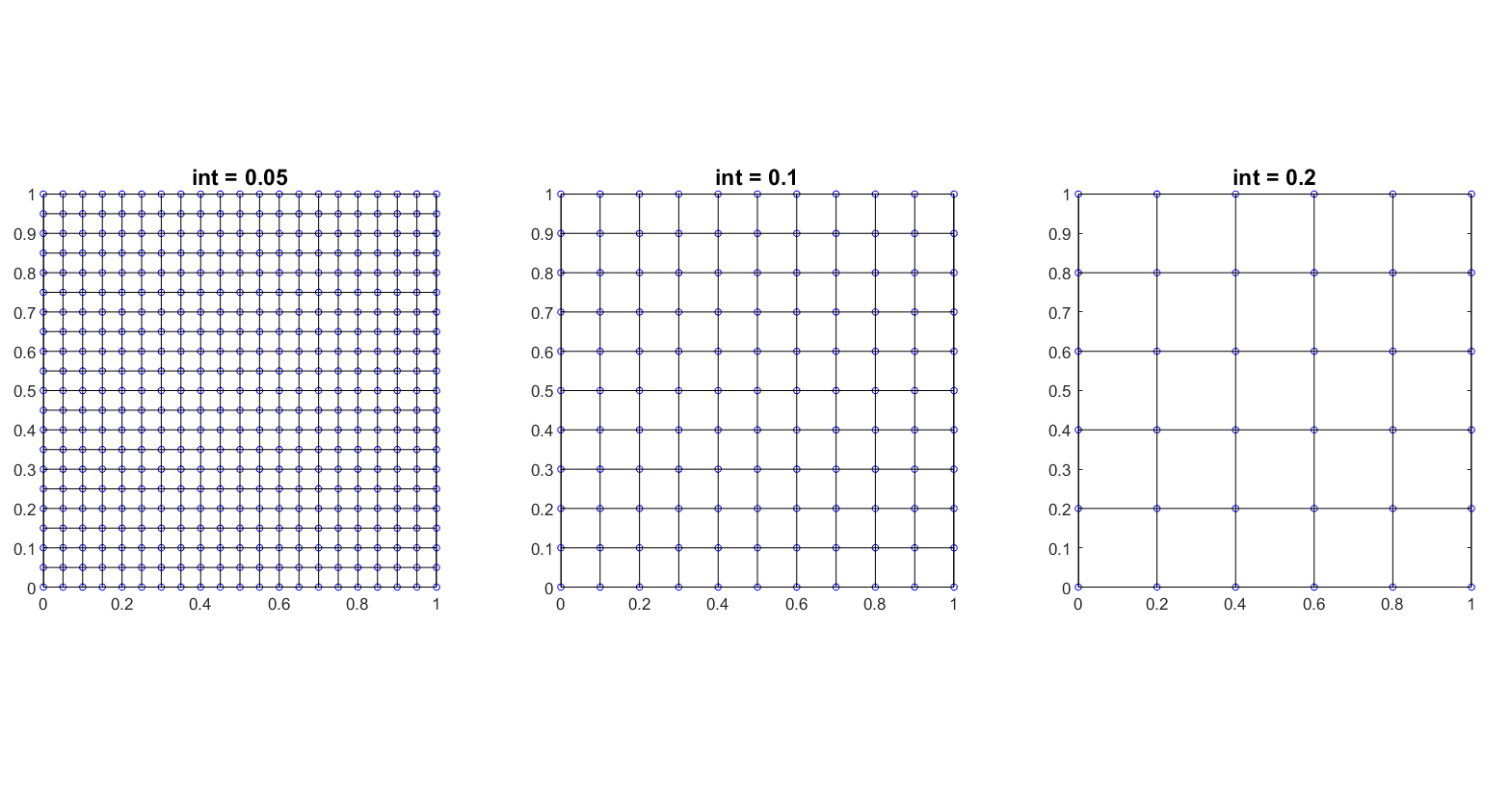
Methods for graph creation from point clouds
In my Graph-Creation-from-point-clouds repository I have included four function files which create graphs according to different rules. Each function takes a set of points (nodes) in a space of arbitrary dimensionality, and decides whether each pair of nodes is connected. The functions output the network in the form of an edgelist: each row is one edge [node i, node j, distance between].
See Compare_graph_creation_functions.m for a script to create the figures below.
K nearest-neighbors Graph
Create_KNN_graph.m
Connect each node to its k nearest neighbors. [node i, node j, distance] is included in edgelist if j is a nearest neighbor of i (the reverse may or may not be true). This type of graph creation rule produces directed edges due to the asymmetry of KNN.
 Using K=3
Using K=3
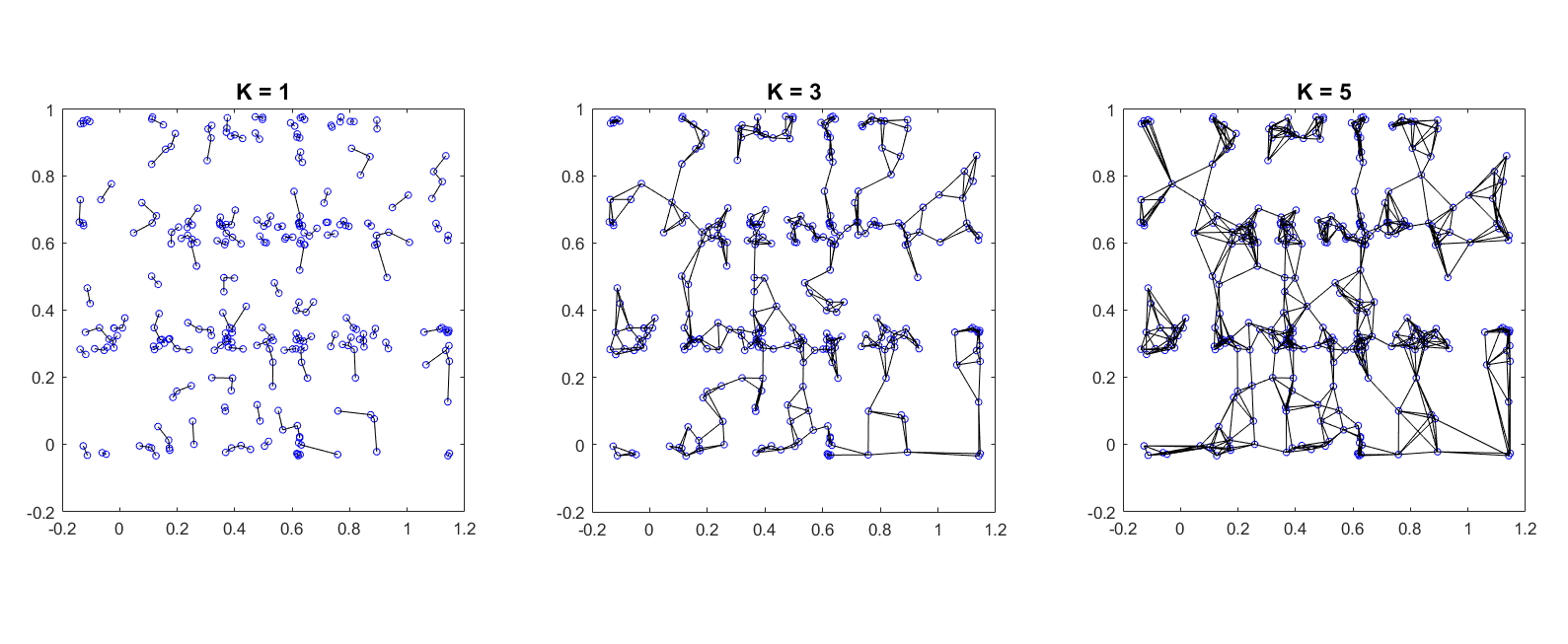 Varying K
Varying K
Epsilon ball Graph
Create_epsilonball_graph.m
Connect each node to all others within a distance \(\epsilon\). This graph will be undirected.
Fixed \(\epsilon\)
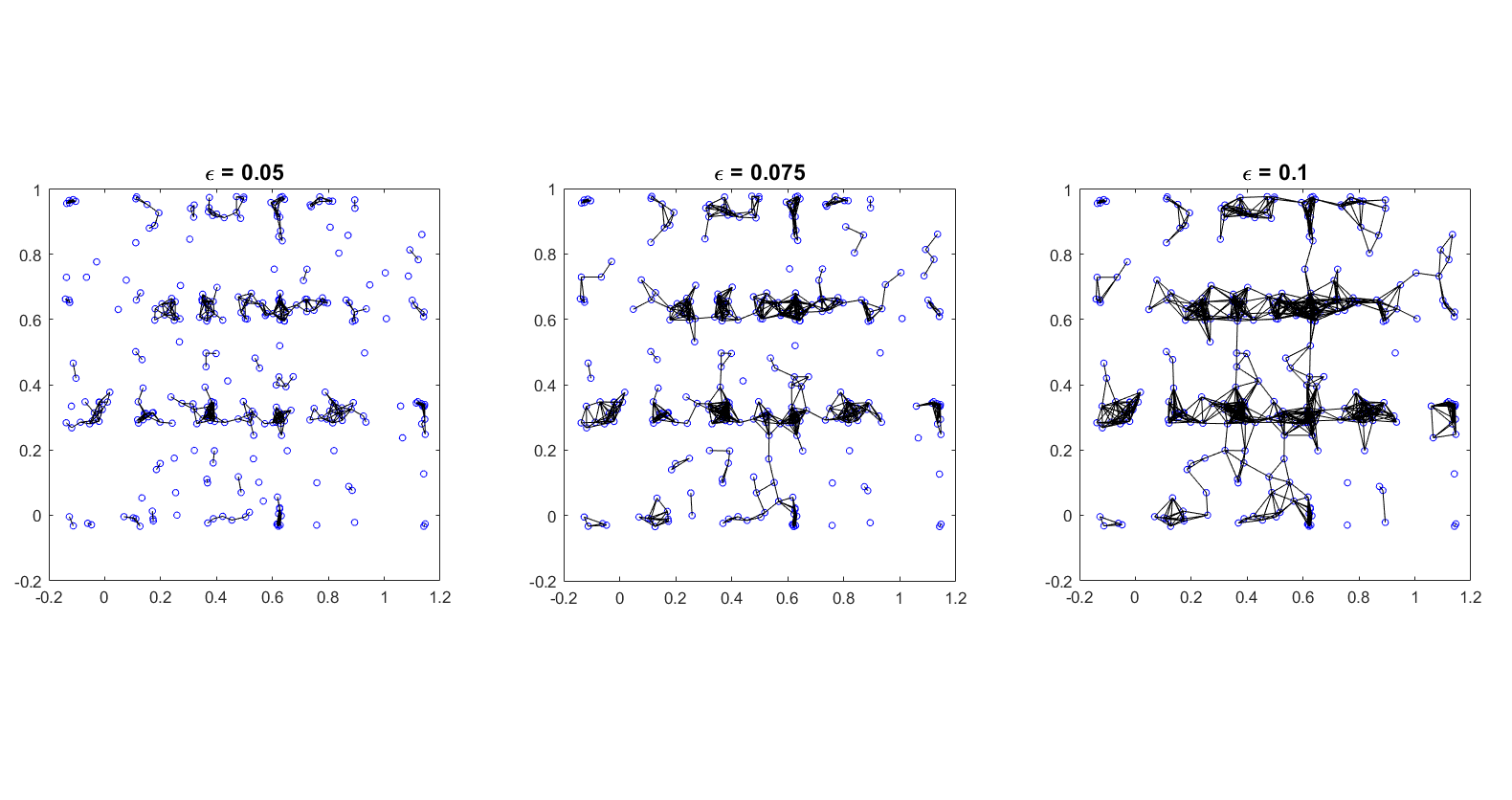
Dynamic \(\epsilon\)
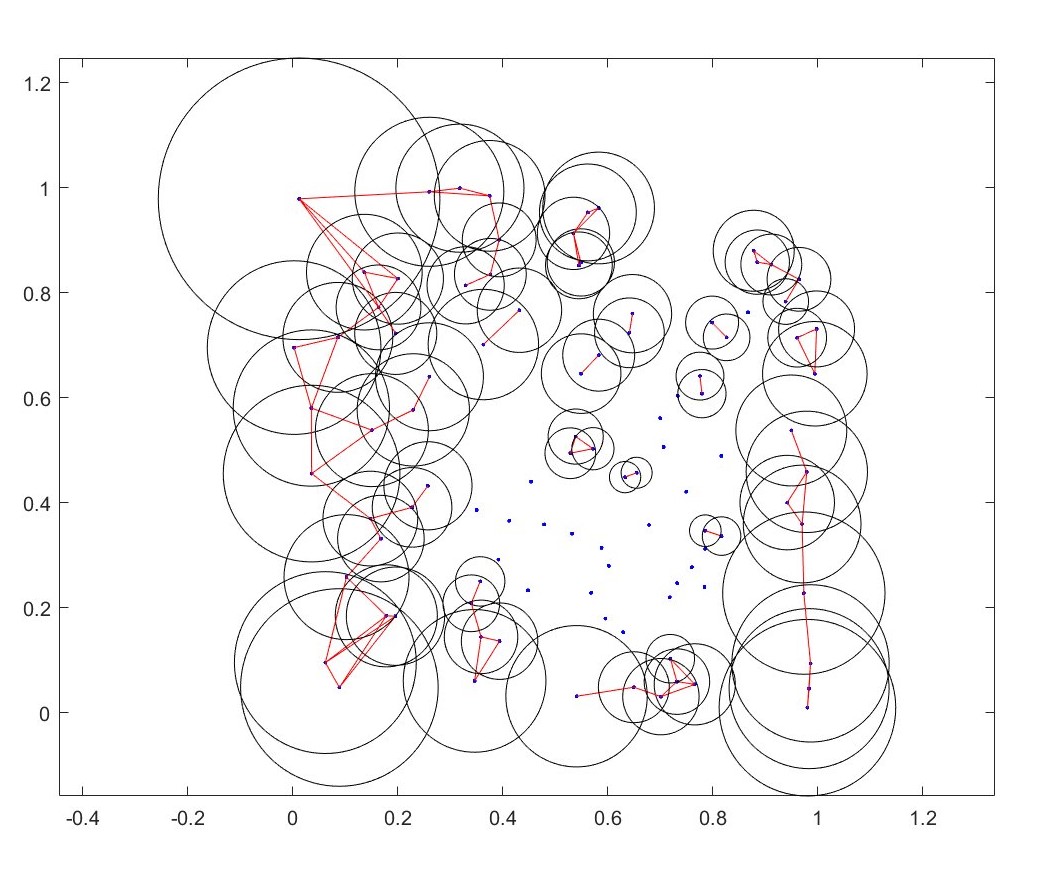 You can also make the epsilon radius epsilon radius a function of the data itself. Above I’ve made \(\epsilon\) inversely proportional to a metric of the local density of points. Each point now uses its own \(\epsilon\) circles). Local density was measured similarly to using a radial basis kernel (gaussian if youre in physics; normal distribution if youre in statistics..) but instead uses a purely exponentially decaying weight.
You can also make the epsilon radius epsilon radius a function of the data itself. Above I’ve made \(\epsilon\) inversely proportional to a metric of the local density of points. Each point now uses its own \(\epsilon\) circles). Local density was measured similarly to using a radial basis kernel (gaussian if youre in physics; normal distribution if youre in statistics..) but instead uses a purely exponentially decaying weight.
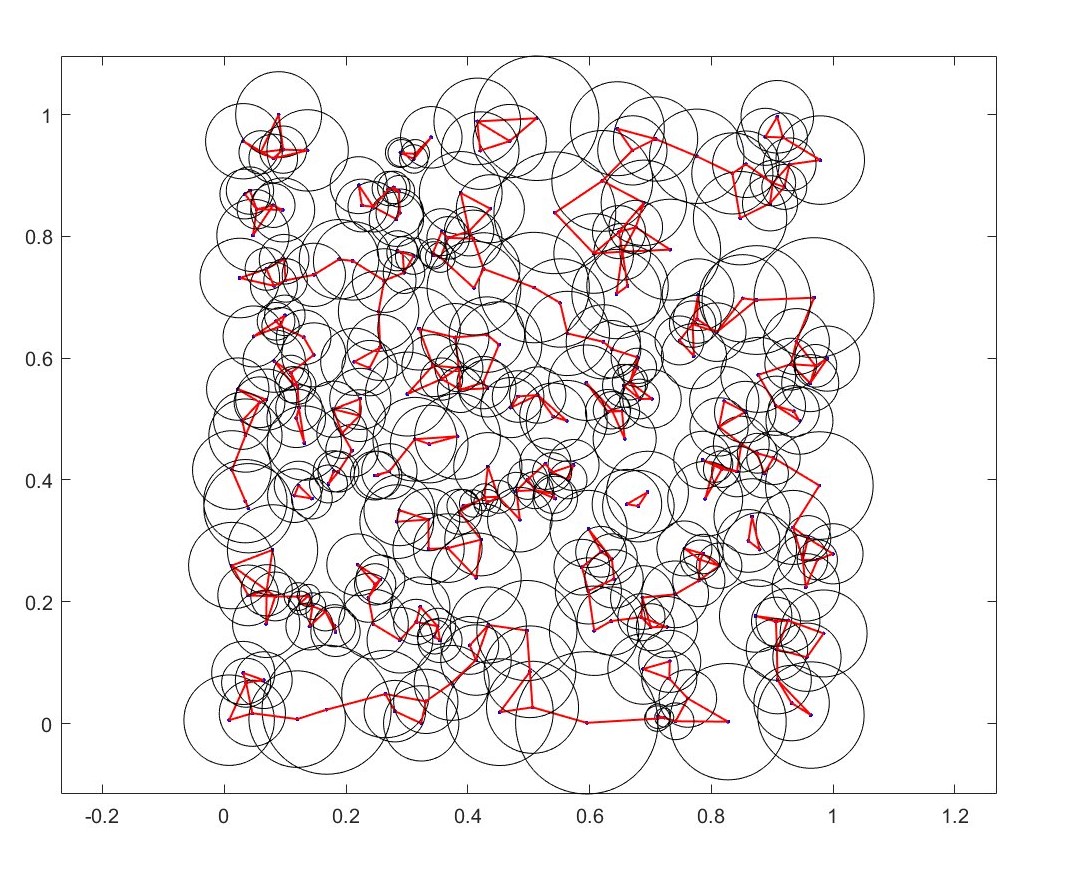 Here \(\epsilon\) radius is proportional to the mean of the distance to second and third nearest neighbors.
Here \(\epsilon\) radius is proportional to the mean of the distance to second and third nearest neighbors.
Tau Rule Graph
Create_Tau_Rule_graph.m
This graph decides whether each pair of nodes are connected according to the tau rule. As of now I cant find the source where I learned about this.
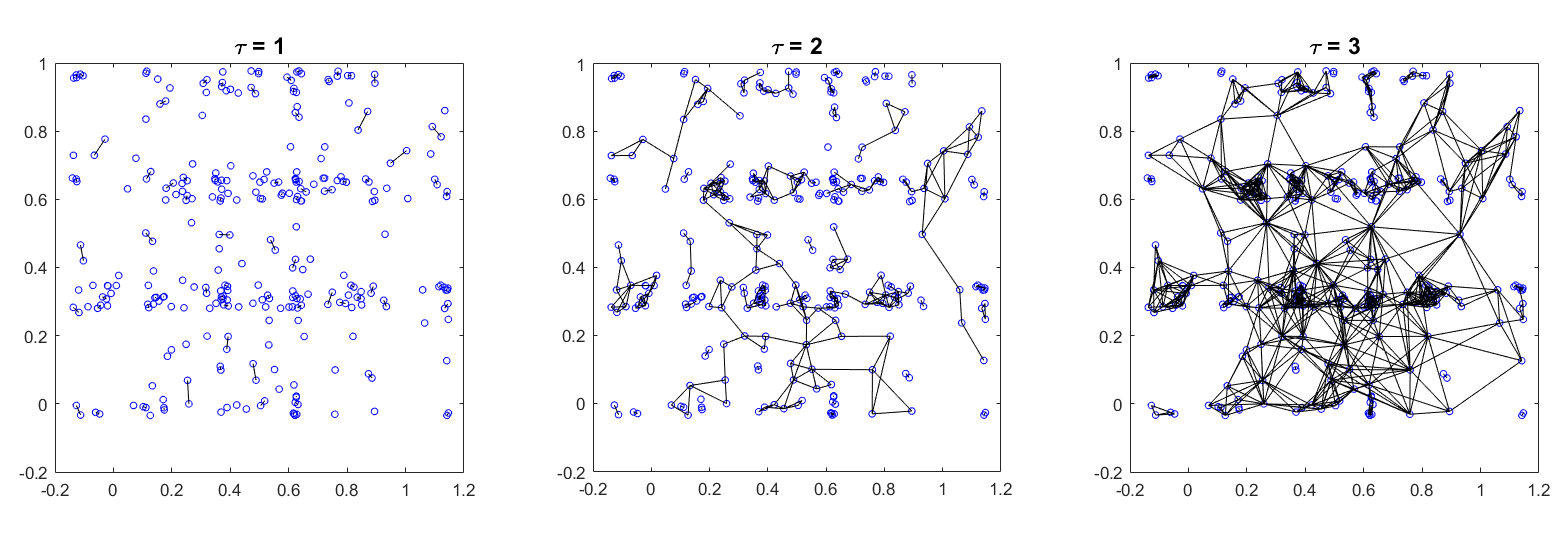 The same data set using different \(\tau =2.2\) values.
The same data set using different \(\tau =2.2\) values.
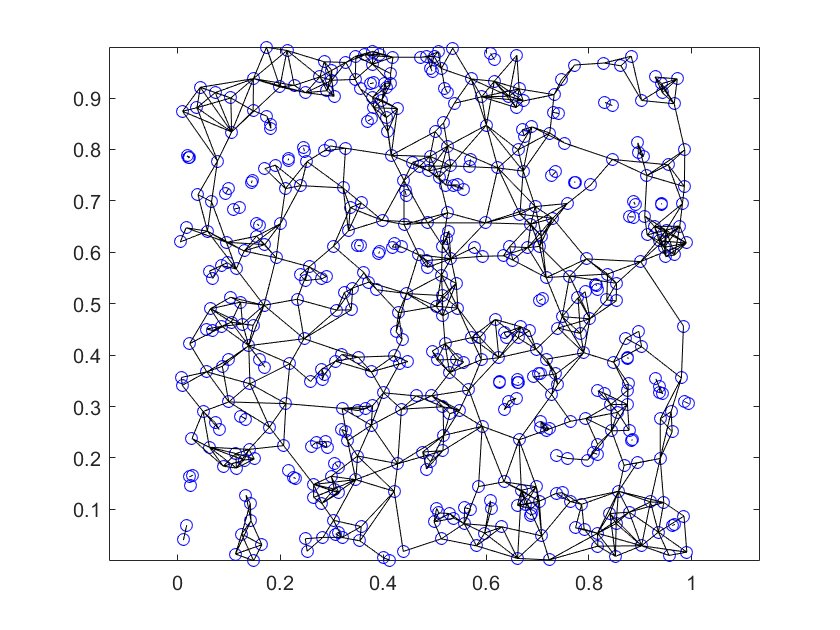 \(\tau =2.2\) with 500 points.
\(\tau =2.2\) with 500 points.
function [edglst]=Create_Tau_Rule_graph(dat,tau)
% builds graph from data using tau rule
%INPUT: dat=data matrix [n x d] : n points in R^d
%tau = positive number >= 1
%OUTPUT: UNDIRECTED edge list [#edges x 3]. d is the distance
%(used as edge weight) along that edge
K=1;%do not alter!
[N, dist]=knnsearch(dat,dat,'K',K+1,'NSMethod','kdtree','Distance','Euclidean');%NOTE: nearest neighbor sets are not necessarily symmetric
N=N(:,2:end); %index of each point's nearest neighbor. first col is each point itself
dist=dist(:,2:end); %distance to each point's nearest neighbor
edglst=[];
M=size(dat,1);
for u=2:M
for v=1:u-1
if (dist(u)<=tau*dist(v))&&(dist(v)<=tau*dist(u))
if (norm(dat(u,:)-dat(v,:))<=tau*dist(u))||(norm(dat(u,:)-dat(v,:))<=tau*dist(v))
edglst=[edglst; u, v, norm(dat(u,:)-dat(v,:))];
end
end
end
end
end
Create 2D rectangular lattice graph
The function Create_Lattice_Network_2D.m allows you to obtain the set of edges connecting points in a rectangular lattice.